December 2016
ENIGMA-Russia
Neuroscience in Pediatrics, Scientific Center of Children’s Health, December 12, 2016
Big Data & the Brain, Skolkovo Institute of Science and Technology, December 13, 2016
ENIGMA at SIPAIM
Congratulations to the 9 talks awarded to SIPAIM this year.
October 2016
10/27/2016 ENIGMA at MICCAI
ENIGMA at MICCAI
In addition to the awardees described below, members from the ENIGMA Consortium’s USC site presented the following papers and satellite symposium at MICCAI in Athens, Greece this month. A Continuous Model of Cortical Connectivity An Empirical Study of Continuous Connectivity Degree Sequence Equivalents Using multiple Diffusion MRI measures to predict Alzheimer’s Disease with a TV-L1 prior A volumetric conformal mapping approach to spectral clustering of white matter fibers Multi-modal Registration Improves Group Discrimination in Pediatric Traumatic Brain Injury
10/20/2016Awards at MICCAI

Major congratulations to ENIGMA local (USC) Dan Moyer for his young scientist award-winning talk at MICCAI. See details here.
10/03/2016Nature Neuroscience publishes ENIGMA-CHARGE Consortia’s collaboration on genome-wide association study of intracranial brain volume
In the largest study of its kind, the ENIGMA consortium paired up with the CHARGE consortium for a GWAS of intracranial brain. Full article is available through Nature Neuroscience and here.
09/09/2016Subcortical Brain Volume in OCD: American Journal of Psychiatry
See full article here.
June 2016
06/07/2016The ENIGMA Interviews: Neuroscientists Discuss Worldwide Successes, Challenges
ENIGMA has interviewed 13 world-renowned brain scientists, on their recent successes in discovering what helps or protects our brains, by pooling 53,000 brain scans and DNA from 35 countries worldwide.
As leaders of some of ENIGMA’s 30 working groups, the scientists explain how they are organizing the world’s largest neuroimaging studies of depression, addiction, schizophrenia, bipolar illness, obsessive compulsive disorder, among the 18 brain diseases that ENIGMA studies. They discuss the challenges and successes in integrating data collected from medical centers worldwide, and some of the benefits of working in very large global teams to study factors that help or harm the human brain. ENIGMA’s ongoing studies have brought together the talents and expertise of leaders from 35 countries. Read part 1, from Atlanta, using the link below: https://goo.gl/fu2NYT (100MB, .pdf) Part 2, from Geneva, to come later this month!
Don’t forget to follow ENIGMA on Facebook! Click the link below:
06/01/2016[Follow ENIGMA on Facebook]
This June in Geneva, ENIGMA scientists will meet collaborators from all over the world and give over 30 presentations at the Organization for Human Brain Mapping. Abstracts of these studies are now searchable online (text search: “ENIGMA”).
May 2016
05/21/2016Hong Kong, Atlanta events showcase ENIGMA’s worldwide brain initiatives

 In a single week, ENIGMA researchers gathered in both Atlanta and Hong Kong to present the world’s largest brain imaging studies of major depression, obsessive compulsive disorder, and schizophrenia – and landmark findings from over 18 diseases that ENIGMA now studies. In Atlanta, Dr. Lianne Schmaal, who co-leads ENIGMA’s Major Depression initiative, presented data from over 10,000 individuals, detecting consistent patterns of brain abnormalities in people who are depressed worldwide. The landmark study, published in this month’s issue of Molecular Psychiatry freely available here,brought together experts from 57 institutions – from Australia, North America, Western Europe and Russia. The researchers combed through brain scans, detecting consistent patterns of abnormalities in the frontal and limbic brain regions. These brain regions have long been implicated in depressive symptoms and are the target of several antidepressant treatments. Dr. Odile van den Heuvel, a psychiatrist from the Netherlands who co-leads ENIGMA’s initiatives on obsessive compulsive disorder, also presented data from 26 OCD sites worldwide (including 1830 OCD patients and 1759 controls), revealing distinct patterns of brain abnormalities in children and adults with OCD. ABSTRACT HERE . ENIGMA researchers led a series of 4 symposia, at the Society for Biological Psychiatry in Atlanta, highlighting their large-scale brain studies. Many of them – such as ENIGMA’s Lifespan project presented by Dr. Sophia Frangou ABSTRACT HERE -are the largest brain studies ever performed – in terms of the numbers of scans, and the diversity of participation by scientists worldwide. In Hong Kong, ENIGMA PI Paul Thompson highlighted ENIGMA’s close partnerships with international networks such as AUSSIE, led by neurologist Dr. Jeff Looi, which brings together experts worldwide to study neurodegenerative diseases. At this event, at the Royal Australian and New Zealand Congress on Psychiatry, Dr. Looi noted the unprecedented power of global consortia to detect how treatments resist brain decline or symptoms of brain disease. ENIGMA scientists and collaborators at last week’s events in Hong Kong (left; May 9-10) and Atlanta (right, May 12-14). Pictured: From left to right, in Hong Kong: Christos Pantelis, Dennis Velakoulis, Brian Power, Rosa Molina, Jeff Looi, Mark Walterfang, Benny Liberg, Alexander Santillo, Conor Owens-Walton, Fiona Wilkes, Bjorn Cartledge, Paul Thompson; From left to right, in Atlanta: Barbara Franke, Chris Whelan, Patricia Conrod, Odile van den Heuvel, Ole Andreassen, Sarah Medland, Paul Thompson, Hugh Garavan, Lianne Schmaal, Theo van Erp, Neda Jahanshad, Dick Veltman.
In a single week, ENIGMA researchers gathered in both Atlanta and Hong Kong to present the world’s largest brain imaging studies of major depression, obsessive compulsive disorder, and schizophrenia – and landmark findings from over 18 diseases that ENIGMA now studies. In Atlanta, Dr. Lianne Schmaal, who co-leads ENIGMA’s Major Depression initiative, presented data from over 10,000 individuals, detecting consistent patterns of brain abnormalities in people who are depressed worldwide. The landmark study, published in this month’s issue of Molecular Psychiatry freely available here,brought together experts from 57 institutions – from Australia, North America, Western Europe and Russia. The researchers combed through brain scans, detecting consistent patterns of abnormalities in the frontal and limbic brain regions. These brain regions have long been implicated in depressive symptoms and are the target of several antidepressant treatments. Dr. Odile van den Heuvel, a psychiatrist from the Netherlands who co-leads ENIGMA’s initiatives on obsessive compulsive disorder, also presented data from 26 OCD sites worldwide (including 1830 OCD patients and 1759 controls), revealing distinct patterns of brain abnormalities in children and adults with OCD. ABSTRACT HERE . ENIGMA researchers led a series of 4 symposia, at the Society for Biological Psychiatry in Atlanta, highlighting their large-scale brain studies. Many of them – such as ENIGMA’s Lifespan project presented by Dr. Sophia Frangou ABSTRACT HERE -are the largest brain studies ever performed – in terms of the numbers of scans, and the diversity of participation by scientists worldwide. In Hong Kong, ENIGMA PI Paul Thompson highlighted ENIGMA’s close partnerships with international networks such as AUSSIE, led by neurologist Dr. Jeff Looi, which brings together experts worldwide to study neurodegenerative diseases. At this event, at the Royal Australian and New Zealand Congress on Psychiatry, Dr. Looi noted the unprecedented power of global consortia to detect how treatments resist brain decline or symptoms of brain disease. ENIGMA scientists and collaborators at last week’s events in Hong Kong (left; May 9-10) and Atlanta (right, May 12-14). Pictured: From left to right, in Hong Kong: Christos Pantelis, Dennis Velakoulis, Brian Power, Rosa Molina, Jeff Looi, Mark Walterfang, Benny Liberg, Alexander Santillo, Conor Owens-Walton, Fiona Wilkes, Bjorn Cartledge, Paul Thompson; From left to right, in Atlanta: Barbara Franke, Chris Whelan, Patricia Conrod, Odile van den Heuvel, Ole Andreassen, Sarah Medland, Paul Thompson, Hugh Garavan, Lianne Schmaal, Theo van Erp, Neda Jahanshad, Dick Veltman.
05/02/2016
The Future of ENIGMA, Genomics and the Human Brain – An Interview
 The Russian edition of “Scientific American” magazine features an article on ENIGMA, highlighting worldwide alliances that investigate 15 major diseases of the brain, in the largest neuroimaging studies in history.
The Russian edition of “Scientific American” magazine features an article on ENIGMA, highlighting worldwide alliances that investigate 15 major diseases of the brain, in the largest neuroimaging studies in history.
ENIGMA director Paul Thompson is interviewed, discussing the consortium’s history, goals and recent discoveries. ENIGMA will host a workshop for Russian members in December, in partnership with the Moscow Child Health Center.
The article is available here –
[in ENGLISH]
https://docs.google.com/document/d/1XhJ6tyx-WNJJNzAsjQuYaJDtQAadYl6aKDpsE1GcaZ0/edit#
[and in RUSSIAN]
http://scientificrussia.ru/articles/ne-genom-edinym-zhivet-mozg
April 2016
04/27/2016ENIGMA in Prague
ENIGMA scientists showcased their work in 8 technical papers at this month’s ISBI 2016 conference, collated here: http://bit.ly/1qVBtxz
One paper earned the prestigious ISBI 2016 Best Student Paper Award for first author, Qi Wang: the authors showed how to merge information from numerous sources on brain connectivity to better detect disease, applying novel concepts such as L1 fusion to brain networks and connectivity maps.
Other papers boosted the power to detect brain disease by using new metrics from diffusion tensor images (Nir et al., see booklet), and tensor based morphometry; ENIGMA’s sparse learning algorithms were also showcased for encoding and decoding patterns in large databases of brain images, genomes, and clinical data (Zhu et al., and Lorenzi et al.).
ENIGMA is now performing a very large scale worldwide analysis of factors that affect the cortical patterns of the human brain (see Pizzagalli et al., in the booklet).
04/01/2016
Dana Foundation highlights ENIGMA’s Global Psychiatry Initiatives
A news article from the Dana Foundation (http://www.dana.org/News/Team_Science/)
highlights’ ENIGMA’s growing role in bringing together the scientific community to discover information in data, at a scale beyond the ability of any single lab to gather and analyze. Award-winning science journalist, Carl Sherman, highlights ENIGMA’s initiatives – including a recent partnership with the Psychiatric Genomics Consortium, analyzing brain imaging and genetic data on more than 50,000 people, enlisting more than 500 investigators world-wide to seek connections between mental illness and brain morphology and function. In the latest paper, in Nature Neuroscience, researchers matched genes associated with schizophrenia risk from GWAS of more than 76,000 patients and controls in the PGC dataset against genes associated with brain volume in 11,840 subjects gathered by ENIGMA. the work was a successful proof-of-concept study, says Thompson, “a roadmap of partnership between PGC and ENIGMA. It showed that you can pool genomic and imaging data in a way to test whether psychiatric risk genes alter brain structure or function.” Such a partnership represents “a two-way conversation, between gene-hunters in imaging and in psychiatry, comparing their hands of cards.” – See more at: http://www.dana.org/News/Team_Science/
March 2016
ENIGMA Center members present 6 peer-reviewed papers at the SPIE Medical Imaging Conference, San Diego, CA, USA
 Each new paper presents a novel algorithm for handling “Big Data” of various kinds – from distributed analyses of brain scans from across the world, to new methods in information theory to detect patterns – and see trends – in blood markers of disease. One new article describes how to use “sparse learning” or compressive sensing to create data dictionaries for massive datasets of functional brain maps; another shows how to create and handle the rich new 6-dimensional brain scans called “Q-Space Images” – a new tool to understand brain disease and probe effects of genes and interventions, using phenomenally complex data acquisition protocols. Click here to download the full set of SPIE papers.
Each new paper presents a novel algorithm for handling “Big Data” of various kinds – from distributed analyses of brain scans from across the world, to new methods in information theory to detect patterns – and see trends – in blood markers of disease. One new article describes how to use “sparse learning” or compressive sensing to create data dictionaries for massive datasets of functional brain maps; another shows how to create and handle the rich new 6-dimensional brain scans called “Q-Space Images” – a new tool to understand brain disease and probe effects of genes and interventions, using phenomenally complex data acquisition protocols. Click here to download the full set of SPIE papers.
Image, left: Graph of latent factors for brain MRI biomarkers of cognitive decline constructed by CorEx.
From Madsen et al., SPIE Medical Imaging, 2016.
February 2016
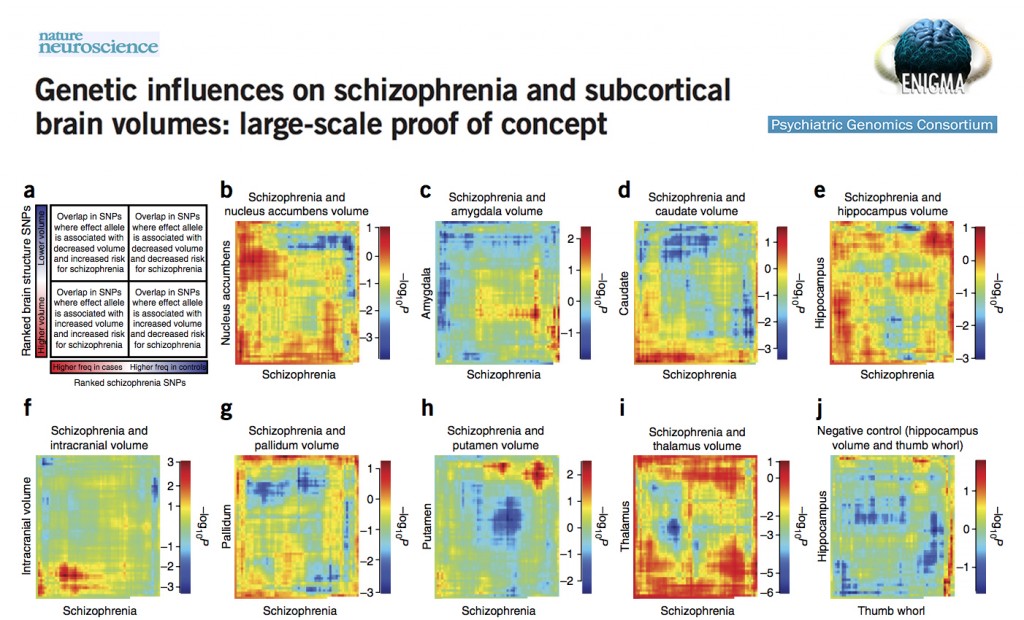
ENIGMA and PGC Publish a Joint Roadmap for Future Collaborations
ENIGMA has partnered with the Psychiatric Genomics Consortium (PGC; http://pgc.unc.edu/) to investigate possible genetic ‘overlap’ between brain measures (e.g. subcortical volume, white matter microstructure) and risk for psychiatric disorders (e.g. schizophrenia, bipolar disorder). The first major collaboration between the consortia, published in this month’s Nature Neuroscience, provides a comprehensive roadmap for future studies of genetic covariance, applying a series of novel techniques (including Rank Rank Hypergeometric Overlap (RRHO), genetic predisposition scoring, conjunction analysis and linkage disequilibrium score regression (LDSR)) to test for possible statistical overlap between a large-scale genome-wide association (GWA) meta-analysis of schizophrenia (PGC, Nature, 2014) and seven independent GWA meta-analyses of subcortical brain volumes, including the nucleus accumbens, amygdala, caudate nucleus, hippocampus, pallidum, putamen and thalamus, in addition to an eighth independent GWA meta-analysis of intracranial volume (ICV; Hibar et al., Nature, 2015). The study did not reveal any significant genetic correlations between subcortical/intracranial volume measures and risk for schizophrenia. However, as the ENIGMA consortium expands its genome-wide association studies of novel brain phenotypes (including cortical thickness/surface area and white matter microstructure) and the PGC explores a broad body of psychiatric illnesses with ever-increasing sample sizes, this inaugural study should serve as an important proof-of-concept for future collaborations between the two. The full paper can be downloaded at the following link: http://www.nature.com/neuro/journal/vaop/ncurrent/full/nn.4228.html
December 2015
Russian Participation in ENIGMA hailed at Moscow Science Week
 Pictured: (from top left): Russian Deputy Minister for Science and Education, Lyudmila Ogorodova, Paul Thompson, Dr Alexander Baranov, Dr Leyla Baranova Namazova; Russian members of ENIGMA; Dr Vladimir Zelman awarding prizes to Skoltech students; Lyubomir Aftanas, ENIGMA member from Novosibirsk, Siberia; Dr Alexander Kuleshov addresses “Moscow Science Week” at the Russian National Academy of Sciences. For more photos, please see: https://plus.google.com/photos/106195383001884599779/albums/6227000582976975601
Pictured: (from top left): Russian Deputy Minister for Science and Education, Lyudmila Ogorodova, Paul Thompson, Dr Alexander Baranov, Dr Leyla Baranova Namazova; Russian members of ENIGMA; Dr Vladimir Zelman awarding prizes to Skoltech students; Lyubomir Aftanas, ENIGMA member from Novosibirsk, Siberia; Dr Alexander Kuleshov addresses “Moscow Science Week” at the Russian National Academy of Sciences. For more photos, please see: https://plus.google.com/photos/106195383001884599779/albums/6227000582976975601
MOSCOW, 12/11/15 In a series of interviews on Russian national television, Dr. Vladimir Zelman, ENIGMA Coordinator in Russia, noted the vibrant partnerships in ENIGMA across Russia that are now producing groundbreaking discoveries about the human brain. The Russian Deputy Minister for Science and Education, Dr. Lyudmila Ogorodova, attended ENIGMA’s meetings and noted her strong support for ENIGMA and its established and extensive partnerships with other Russian science initiatives, such as Co-Brain, and NeuroNEXT. ENIGMA’s 500 scientists now study 12 brain diseases in over from 35 countries, and have published the largest-ever genomic analyses of the brain. As part of a week of ENIGMA-related events in Russia, Dr. Zelman, and ENIGMA director Paul Thompson, noted the remarkable breadth of participation in ENIGMA by mathematicians, geneticists, and clinical neuroscientists across Russia. “We see a great surge of interest in ENIGMA, which is widely known across Russia”, Zelman told Rossiya 24, a national news channel of the Russian Federation. Zelman is a Russian-born scientist, trained at the Meshalkin Institute in Novosibirsk. A member of the Russian National Academy of Sciences, Zelman has helped to build up a network of scientists contributing to ENIGMA across Russia. Russian scientists are now deeply involved in ENIGMA’s projects on Parkinson’s disease, major depression, post traumatic stress, and developmental disorders in children, including ADHD. ENIGMA co-founder, Paul Thompson, co-hosted a planning meeting for Russian ENIGMA members chaired by developmental neuroscientist Dr. Leyla Namazova, at the Child Health Institute in Moscow, on December 8th. The meeting, attended by ENIGMA laboratory directors from across Russia, Siberia, and Ukraine, gave all scientists a forum to update each other on their scientific work as part of ENIGMA. Interviewed by Scientific American’s Russian magazine, Thompson pointed to developments being accelerated by ENIGMA’s Russian scientists in the fields of genomics, machine learning, and analysis of brain connectivity, such as finding patterns in medical data to detect and classify autism spectrum disorder and Alzheimer’s disease. Dr. Namazova, who co-directs Moscow’s largest children’s hospital, chaired an evening session about ENIGMA with institute directors, radiologists and computer scientists from the Moscow Institute of Neurology, Siberian National Academy of Sciences, and Skoltech University (see photo). Paul Thompson supported the Moscow Child Health Center’s plans to host a training workshop for ENIGMA next December in Moscow – training young scientists is a key mission of ENIGMA, which now supports a productive international exchange program. Zelman hosted an awards ceremony for young scientists at Skoltech – Moscow’s $100M technical university focusing on engineering and business start-ups in informatics – after Thompson’s keynote talk highlighting ENIGMA’s work in Russia. Sergey Illarioshkin, MD, a leader in the genetics of Parkinson’s disease, presented his team’s work at the Moscow Institute of Neurology, mapping brain abnormalities as part of ENIGMA’s Parkinson’s disease working group. This group now analyzes brain MRI scans of Parkinson’s disease patients from Russia, Italy, and The Netherlands, to identify preventable risk factors for the disease. Lyubomir Aftanas MD PhD, a psychiatrist who runs a brain mapping center in Novosibirsk, presented a his work on ENIGMA-Depression, which compares Siberian datasets to others worldwide, in what is now the largest brain imaging study of depression ever conducted (see photo). “ENIGMA gives us a power we have not had”, said Aftanas. “We contribute data and expertise to ENIGMA’s efforts in neurology and psychiatry; after brainstorming in Moscow this week, we are keen to join ENIGMA’s epigenetics and lifespan projects, among others, to study modifications of the genome throughout life and how they impact the brain. In Siberia, we are collecting unique data to help answer these questions”. At the meeting, neurosurgical delegates from Ukraine outlined their plans to participate in ENIGMA next year, offering to contribute genetic and brain scan data from patients with a range of brain disorders. In two days of highly animated sessions of the Russian National Academy of Sciences, a series of workshops and symposia hailed ENIGMA’s work bringing scientists together, as part of “Moscow Science Week”. ENIGMA director Paul Thompson thanked senior members of the Russian Academy, including its President, noted physicist Vladimir Fortov, for supporting ENIGMA and promoting its ongoing projects to their colleagues; Karkhevich Institute director, Dr. Alexander Kuleshov, noted that he is sending 3 mathematicians to the ENIGMA Center in Los Angeles to boost the ongoing machine learning efforts to discover genomic markers of brain disease. Kuleshov leads “big data” analysis efforts with Airbus and aerospace industry partners. He pointed to the power of worldwide collaboration in informatics in solving pressing problems in science. Using large-scale analyses, ENIGMA has been publishing “big data” papers with hundreds of scientists in high-impact journals such as Nature and Nature Genetics. Russian scientists have long been a key source of expertise for ENIGMA, since noted Moscow geneticist Kazima Bulayeva published a series of reports in 2012 from her cohorts in the highlands of Dagestan. Her work reveals how genes implicated by ENIGMA affect brain structure and people’s risk for mental retardation and psychosis. Understanding the transmission of these genetic sources of risk underlies partnerships between her team’s work in populaton genetics and ENIGMA’s work on the genetics of the human brain. Interviewed by Russia’s Scientific American, Paul Thompson was asked if ENIGMA’s mathematicians had found what the Russian poet Pushkin described as “harmony in the brain”, pointing to ENIGMA’s findings mapping the brain’s interacting circuits and systems. Vladimir Zelman pointed to both successes and challenges leading ENIGMA’s efforts in Russia, and how Russian politicians and institute directors across Moscow and Siberia have helped to overcome them. The full interview will be published in Russia’s Scientific American, next month.
ENIGMA and the Individual – An Up-to-Date Review of ENIGMA
12/09/2015 ENIGMA has published a broad overview of its activities in the December 2015 issue of the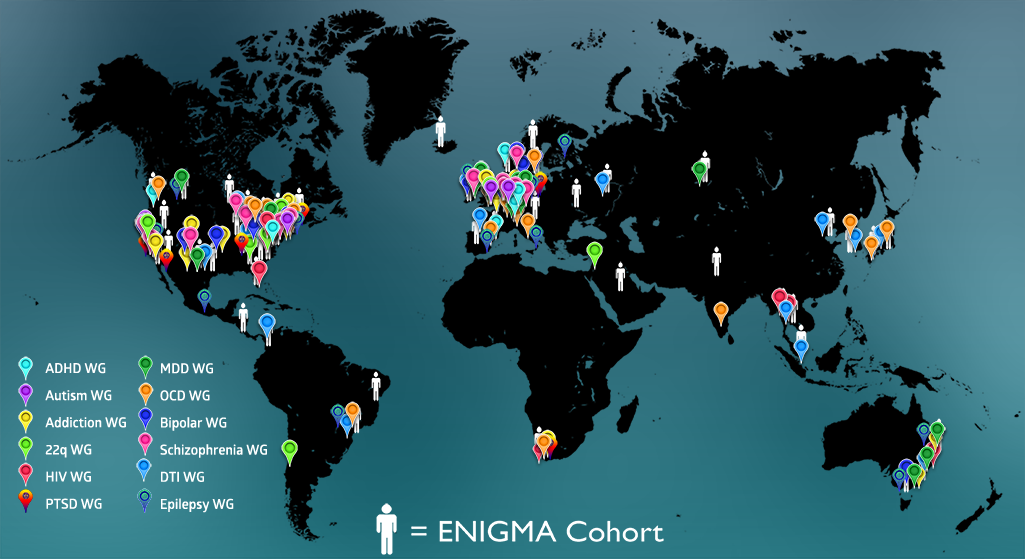 journal, NeuroImage. In the review, available here, we discuss recent work by the ENIGMA Consortium (http://enigma.ini.usc.edu) – a global alliance of over 500 scientists spread across 200 institutions in 35 countries collectively analyzing brain imaging, clinical, and genetic data. Initially formed to detect genetic influences on brain measures, ENIGMA has grown to over 30 working groups studying 12 major brain diseases by pooling and comparing brain data. In some of the largest neuroimaging studies to date – of schizophrenia and major depression – ENIGMA has found replicable disease effects on the brain that are consistent worldwide, as well as factors that modulate disease effects. In partnership with other consortia including ADNI, CHARGE, IMAGEN and others, ENIGMA’s genomic screens – now numbering over 30,000 MRI scans – have revealed at least 8 genetic loci that affect brain volumes. Downstream of gene findings, ENIGMA has revealed how these individual variants – and genetic variants in general – may affect both the brain and risk for a range of diseases. The ENIGMA consortium is discovering factors that consistently affect brain structure and function that will serve as future predictors linking individual brain scans and genomic data. It is generating vast pools of normative data on brain measures – from tens of thousands of people – that may help detect deviations from normal development or aging in specific groups of subjects. We discuss challenges and opportunities in applying these predictors to individual subjects and new cohorts, as well as lessons we have learned in ENIGMA’s efforts so far.
journal, NeuroImage. In the review, available here, we discuss recent work by the ENIGMA Consortium (http://enigma.ini.usc.edu) – a global alliance of over 500 scientists spread across 200 institutions in 35 countries collectively analyzing brain imaging, clinical, and genetic data. Initially formed to detect genetic influences on brain measures, ENIGMA has grown to over 30 working groups studying 12 major brain diseases by pooling and comparing brain data. In some of the largest neuroimaging studies to date – of schizophrenia and major depression – ENIGMA has found replicable disease effects on the brain that are consistent worldwide, as well as factors that modulate disease effects. In partnership with other consortia including ADNI, CHARGE, IMAGEN and others, ENIGMA’s genomic screens – now numbering over 30,000 MRI scans – have revealed at least 8 genetic loci that affect brain volumes. Downstream of gene findings, ENIGMA has revealed how these individual variants – and genetic variants in general – may affect both the brain and risk for a range of diseases. The ENIGMA consortium is discovering factors that consistently affect brain structure and function that will serve as future predictors linking individual brain scans and genomic data. It is generating vast pools of normative data on brain measures – from tens of thousands of people – that may help detect deviations from normal development or aging in specific groups of subjects. We discuss challenges and opportunities in applying these predictors to individual subjects and new cohorts, as well as lessons we have learned in ENIGMA’s efforts so far.
November 2015
ENIGMA at SIPAIM, Ecuador
11/27/2015 ENIGMA members and collaborators Dr Emily Dennis, Prof Paul  Thompson, Dan Moyer, and Prof Natasha Lepore gave a series of tutorial lectures in Cuenca, Ecuador, describing new algorithms for large-scale analysis of brain networks computed by ENIGMA members across the world. In conjunction with the SIPAIM meeting in November 2015, the ENIGMA team presented a series of keynote and tutorial lectures on ENIGMA, how to join, and how to deal with the complexity of brain network data in clinical studies, using powerful new mathematical analyses. Topics ranged from generative models of the connectome, to q-space imaging (which yields 6-dimensional data on brain connections), to analyzing data on brain trauma from sites across the world. Many new and longstanding ENIGMA contributors took part in the event, from across South America and internationally.
Thompson, Dan Moyer, and Prof Natasha Lepore gave a series of tutorial lectures in Cuenca, Ecuador, describing new algorithms for large-scale analysis of brain networks computed by ENIGMA members across the world. In conjunction with the SIPAIM meeting in November 2015, the ENIGMA team presented a series of keynote and tutorial lectures on ENIGMA, how to join, and how to deal with the complexity of brain network data in clinical studies, using powerful new mathematical analyses. Topics ranged from generative models of the connectome, to q-space imaging (which yields 6-dimensional data on brain connections), to analyzing data on brain trauma from sites across the world. Many new and longstanding ENIGMA contributors took part in the event, from across South America and internationally. 
October 2015
ENIGMA Center’s Big Data Algorithms MICCAI Conference in Germny
10/28/2015

ENIGMA’s new tools for analyzing large-scale imaging and genomics data were showcased at a range of workshops, from October 5-9, at this year’s MICCAI conference in Munich.
Among the innovative approaches presented were a new method for brain-wide genome-wide screening of massive imaging databases (Jahanshad et al., MICCAIenetic overlap and boost power in imaging genetics (Hibar et al., MICCAI, 2015), and a new concept, called “genetic connectivity”, to detect and model overlap in the results of large-scale genome-wide association studies (Rinker et al., MICCAI, 2015).
Download the full set of papers from the ENIGMA Center here: http://bit.ly/1WiCMol
ENIGMA Co-Founder Dr Sarah Medland receives Theodore Reich Award
10/20/2015
ENIGMA Co-Founder Dr Sarah Medland is the recipient of the 2015 Theodore Reich Award, for innovative work in psychiatric genetics. Announced at the World Congress on Psychiatric Genetics this weekend, in Toronto, this prestigious award honors one scientist per year for exceptional work with a global impact in the field.

Dr Medland is best known for her work as chair of the Genetics Core of the ENIGMA neuroimaging genetics consortium, which analyzes patterns of brain disease worldwide. Over 500 scientists are involved in the work, studying genetic variants and other factors influencing the brain in healthy and diseased populations from 35 countries worldwide. Dr. Medland was the senior author on the largest genomic study of the brain to date, published in Nature earlier this year.
Dr. Medland is a statistical geneticist working on mental health and neurological traits. She is the head of the Quantitative Genetics team at the QIMR Berghofer Medical Research Institute in Brisbane, Australia. The award is named for Dr Theodore (Ted) Reich (1938 – 2003) – first President of the International Society for Psychiatric Genetics, who was an outstanding researcher and mentor to young scientists. The award is made for exceptional published work in psychiatric genetics by candidates who are 40 years or younger in the year of their nomination.
July 2015
ENIGMA Partners to merge techniques from Big Data powerhouses
07/22/2015
 The ENIGMA Center of Excellence, based at the University of Southern California, will work with the KnowEnG (Knowledge Engine for Genomics) Center, based at the University of Illinois, as part of an ambitious new collaboration funded by the Big Data to Knowledge (BD2K) initiative.
The ENIGMA Center of Excellence, based at the University of Southern California, will work with the KnowEnG (Knowledge Engine for Genomics) Center, based at the University of Illinois, as part of an ambitious new collaboration funded by the Big Data to Knowledge (BD2K) initiative.
Drawing upon recent advances in genome sequencing, genome-wide association studies (GWAS) have have led to a wealth of discoveries by mining the human genome for common variants in our genetic sequence that relate to our risk for a range of diseases. A major challenge for GWAS, however, is that the number of SNPs – locations in DNA where people differ in their genetic make-up – is much larger than the number of subjects who can be assessed in a study.
As part of this collaboration, the ENIGMA and KnowEnG centers will mine existing databases of biological pathways and genetic perturbation screening datasets to discover important new genetic interactions and construct networks of gene interactions, called “epistasis networks”. These novel epistasis networks will provide prior information needed to expand sparse learning models and enhance the power of future genome-wide genetic analyses.
This collaborative project will leverage the strengths of the KnowEng center (knowledge of gene-gene interactions) with those of the ENIGMA Center (ongoing development of sparse learning models) to improve our ability to identify causal single nucleotide polymorphisms (SNPs). The project will be coordinated by Jieping Ye and Paul Thompson (ENIGMA) and David Zhao, Jiawei Han & Jun S. Song (KnowEnG).
ENIGMA in Molecular Psychiatry
07/02/2015
Work from the ENIGMA-Major Depressive Disorder and ENIGMA-Schizophrenia working groups has been published in Molecular Psychiatry. The first study, ‘Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium‘, identified (i) significant volume reductions in the hippocampus, amygdala, thalamus, accumbens and intracranial area and (ii) significant volume augmentations in the pallidum and lateral ventricles. The latter volume augmentations were positively associated with duration of illness. The second study, ‘Subcortical brain alterations in major depressive disorder: findings from the Disorder working group‘ , found consistent patterns of hippocampal volume reduction in depressed patients compared to healthy controls. This study, the largest of its kind in MDD, has received press coverage from The Guardian, The Conversation and many other media outlets.
June 2015
ENIGMA at OHBM 2015
06/11/2015Sixteen ENIGMA working groups will present their findings at the 2015 Organisation for Human Brain Mapping meeting. Download our abstracts booklet by clicking on the image below! 
April 2015
ENIGMA in Nature
04.09.2015Our latest GWAS is now out in Nature!  30,000 individual MRIs and genomes from 33 countries — a collaborative effort from 190 institutions Hibar, DP and 300+ co-authors. Common genetic variants influence human subcortical brain structures. Nature, 2015
30,000 individual MRIs and genomes from 33 countries — a collaborative effort from 190 institutions Hibar, DP and 300+ co-authors. Common genetic variants influence human subcortical brain structures. Nature, 2015
March 2015
ENIGMA-Epilepsy
03.09.2015ENIGMA has a new disease working group! ENIGMA-Epilepsy was officially established at The Royal Society, London on Monday, March 9th. Over 20 groups from North and South America, Europe and Australia have already signed up for the initiative. The project is lead by Dr. Christopher D. Whelan (IGC, USC) and Professor Sanjay Sisodiya (UCL). More information can be found on the ENIGMA-Epilepsy webpage.
February 2015
ENIGMA GCTA
02.01.2015Exciting news- ENIGMA is performing Genome-Wide Complex Trait Analysis (GCTA). The effort is lead by Roberto Toro and JB Poline with methodological support from Tom Nichols, with many ENIGMA working groups already involved. Click the GCTA link for more information!
January 2015
Job Openings
01.01.2015We’re hiring! Check out our job openings here
Imaging Genetics Center, Marina Del Rey, California
2014: The ENIGMA Consortium in Review
A retrospective of ENIGMA’s work, spanning the years 2012 to 2014, is now available from Brain Imaging and Behavior. Check it out at the link below!  Thompson, PM and 300+ co-authors. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging and Behavior. SI: Genetic neuroimaging in aging and age-related diseases. DOI 10.1007/s11682-013-9269-5 A list of all ENIGMA publications can also be found at the below link:
Thompson, PM and 300+ co-authors. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging and Behavior. SI: Genetic neuroimaging in aging and age-related diseases. DOI 10.1007/s11682-013-9269-5 A list of all ENIGMA publications can also be found at the below link: